INTRODUCTION
In the treatment of malignant tumors, the identification of therapeutically relevant molecular changes in tumor cells and their interaction with the immune system are increasingly important.1,2 Before new “targeted” agents are used, it is necessary to identify so-called oncogenic driver alterations, i.e. genetic changes that are key to the initiation/spread of the particular malignancy.2 Importantly, similar driver alterations may be present in histologically diverse tumors of different organ systems.2 However, treatment with targeted agents can lead to varying response rates in different tumor types ‒ despite matching gene alterations. For example, in hairy cell leukemia with BRAF V600 mutation, response rates of 96–100% to BRAF inhibitors are described,3 whereas a response rate of 48% has been found in BRAF V600-mutated melanomas.4 In a study of colorectal carcinoma patients with BRAF V600 mutation, no patient responded to targeted monotherapy with vemurafenib.5
Against this background, there has been an increase in the last decade in the use of targeted therapies in phase 1/2 clinical trials in which molecular test results represent an important decision-making criterion for patient inclusion. Molecular changes, rather than the histological characteristics of tumors are relevant for inclusion, and this approach is therefore referred to as “tumor-agnostic” (the term “agnostic” comes from the ancient Greek “agnostikistís”, which means “without knowledge”).6
AUTHORIZED TUMOR-AGNOSTIC AGENTS
In the USA, the first tumor-agnostic approval was granted in May 2017 for the immune checkpoint inhibitor pembrolizumab (Keytruda®), for the treatment of solid tumors in which high microsatellite instability or DNA mismatch repair deficiency is detectable ‒ regardless of the site and histology of the primary tumor.7 On 10 December 2020, the European Medicines Agency’s (EMA’s) Committee for Medicinal Products for Human Use (CHMP) adopted a positive opinion recommending a change to the terms of the marketing authorization for the medicinal product pembrolizumab (Keytruda®).8
In the European Union (EU),9 the first tumor-agnostic agent was authorized in September 2019 with larotrectinib (Vitrakvi®).10 This was followed in May 2020 by its authorization in Switzerland for the treatment of adult and pediatric patients with solid tumors displaying NTRK gene fusions.11,12 Larotrectinib acts as an inhibitor of tropomyosin receptor kinases (TRK) that are encoded by the NTRK genes, and is capable of inhibiting the oncogenic activity of fusion proteins, which can form as a result of fusion of the NTRK gene with other genes in various malignancies (for details, see below).12 According to the label, larotrectinib should only be used if no other satisfactory treatment options are available or if these treatment options have been exhausted. In the clinical studies with larotrectinib, it was at the physician’s discretion whether they considered an available therapy to be “not satisfactory”, meaning whether a patient was unlikely to tolerate or derive clinically relevant benefit from available standard treatments.12 Recently, a prospective, non-interventional phase IV trial with larotrectinib has been launched in different countries worldwide, including Switzerland, to collect further data on safety and efficacy of the drug during routine use (NCT04142437, EUPAS32136).
Another tumor-agnostic agent, entrectinib (Rozlytrek®) was authorized in the USA in August 2019 and in the EU in August 2020 as a ROS1 inhibitor. It was also authorized for the treatment of adult and pediatric patients aged 12 years and above with solid tumors displaying an NTRK gene fusion.13,14 In Switzerland, entrectinib was authorized in December 2020 as a TRK inhibitor (similar to larotrectinib) and as a ROS1 inhibitor.15
MALIGNANT TUMORS WITH NTRK GENE FUSIONS
The TRK receptors TRKA, TRKB, and TRKC are a family of tyrosine kinases that are encoded by the genes NTRK1, NTRK2, and NTRK3, respectively, and play a key role in the development of the nervous system.16,17 NTRK gene fusions with other genes can be detected in many tumors, with more than 80 different fusion partners of NTRK1, NTRK2, and NTRK3 genes.16 With their constitutive activity, the products of these fusion genes can thus act as oncogenic drivers.1,16 For example, a chromosomal translocation, t(12;15)(p13;q25), with the resulting fusion gene ETV6-NTRK3, is detectable in up to 95% of patients with rare tumor types, i.e. infantile fibrosarcomas, congenital mesoblastic nephroma (cellular subtype), secretory breast carcinoma and secretory salivary gland carcinoma.1,16 The constitutively activated fusion proteins stimulate downstream signaling pathways.17 NTRK gene fusions also occur in common tumor types, but at a low frequency.16,17 These more common tumor types include gastrointestinal stromal tumors (GIST) and thyroid cancer (5‒25% NTRK gene fusions in each case), as well as lung, breast, colorectal, pancreatic, renal cell and bile duct carcinomas, melanomas and sarcomas (<5% NTRK gene fusions in each case).17 In the pediatric population, NTRK gene fusions can be found in 4% of diffuse intrinsic pontine gliomas and in 40% of non-brainstem high grade gliomas (infants <3 years).18 An overview of the frequency of NTRK gene fusions in adult and pediatric tumor types is provided in Figure 1.
METHODS FOR DETECTING NTRK GENE FUSIONS
The following established test methods are mainly used for the detection of NTRK gene fusions, which is the necessary prerequisite for prescribing a TRK inhibitor1,20,21:
IMMUNOHISTOCHEMISTRY (IHC)
TRK immunohistochemistry is based on the detection of TRK receptors in tissue samples with pan-TRK antibodies, which are able to bind to the gene products of NTRK1, NTRK2, and NTRK3. This method indicates the presence of a TRK protein but does not clarify whether a fusion is actually present. IHC can therefore be used as an upstream screening tool for common tumor entities that rarely display NTRK gene fusions.1,20 Positive results must be confirmed as “fusion-positive” by means of additional molecular methods.1,20 Pan-TRK immunohistochemistry has a sensitivity of almost 88% and a specificity of around 81% in detecting NTRK fusions overall.22 However, there are differences between the three receptor variants.22 For fusions involving NTRK1 or NTRK2, IHC has been found to have a very high sensitivity of 96.2% and 100% respectively, whereas the sensitivity of IHC for detecting NTRK3 fusions is lower, at 79.4%.22 The sensitivity and specificity of testing also differs for different tumor types (Table 1). Since immunohistochemical testing was performed in only 388 cases, the data should be interpreted with caution.
FLUORESCENCE IN SITU HYBRIDIZATION (FISH)
For the FISH diagnostic technique, fluorescently marked DNA probes ‒ short, single-stranded DNA fragments ‒ are used which bind specifically to particular DNA regions on chromosomes (hybridization). Those hybridization patterns are then evaluated using a microscope. The method can be used to detect chromosomal breakage events in NTRK1, NTRK2, and NTRK3 genes, although a separate test is necessary for each gene investigated. Interpretation of the results is nevertheless complex and requires expert knowledge.1,20 The FISH diagnostic technique is used primarily as a confirmatory analysis in the high-prevalence setting, i.e. for rare tumors where NTRK gene fusion is very likely.1,20
NEXT-GENERATION SEQUENCING (NGS) METHODS
With the NGS-based method, the nucleotide sequence in a DNA molecule is determined using molecular genetics methods.1,20,21 NTRK gene fusion detection therefore occurs at the nucleic acid level and not at the protein level as with IHC. Exact identification of the genes involved in the fusion and of the fusion variants present is thus possible. The disadvantage of such a DNA-based NGS method, however, is that certain intronic/intergenic fusion breakpoints and difficult-to-read GC-rich regions are not covered.1,21 While the specificity of this method for detecting NTRK fusion genes is almost 100%, the sensitivity is only around 81%.22 An RNA-based NGS method or a combination of DNA /RNA based sequencing methods is therefore preferred, as these are better at detecting fusion transcripts and are thus more sensitive.1,21 With RNA-based NGS, the RNA rather than the DNA of the NTRK gene is converted by means of reverse transcriptase to the corresponding cDNA which is then sequenced. Because the large intronic regions of the NTRK genes have already been removed in the RNA by splicing, the difficulties in sequencing are avoided. Nevertheless, RNA extraction from formalin-fixed paraffin embedded tissues is a difficult process. In Swiss pathology institutes, DNA based NGS assays or combinations of DNA/RNA assays from different manufacturers are used to detect NTRK changes.
The advantages and disadvantages of the individual NGS-based methods as well as their interpretation and communication are of key importance for the test algorithms discussed below. High quality molecular diagnostic testing is a prerequisite for the detection of NTRK gene fusions by means of NGS. For Quality Assurance reasons, the relevant tests should therefore be performed only by suitable, designated pathology institutes/accredited hematology molecular laboratories.23 In Switzerland, such analyses are validated by pathologists specializing in molecular pathology.
DIAGNOSTIC ALGORITHMS FOR VARIOUS CLINICAL SCENARIOS
With the authorization of larotrectinib and other TRK inhibitors, testing strategies must now be developed in Switzerland which allow any NTRK gene fusion present to be reliably detected. Recommendations on molecular NTRK testing are decided by interdisciplinary tumor boards involving pathologists and medical oncologists. The European Society for Medical Oncology (ESMO) has recently published general clinical practice recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research.20,24,25 Consistent with the more general approach adopted by the ESMO, other recommendations have been recently proposed which adopt a specific approach for defined clinical scenarios,1,20 as described below.
DGHO (GERMAN SOCIETY FOR HEMATOLOGY AND MEDICAL ONCOLOGY) ALGORITHM FOR DETECTING NTRK GENE FUSIONS
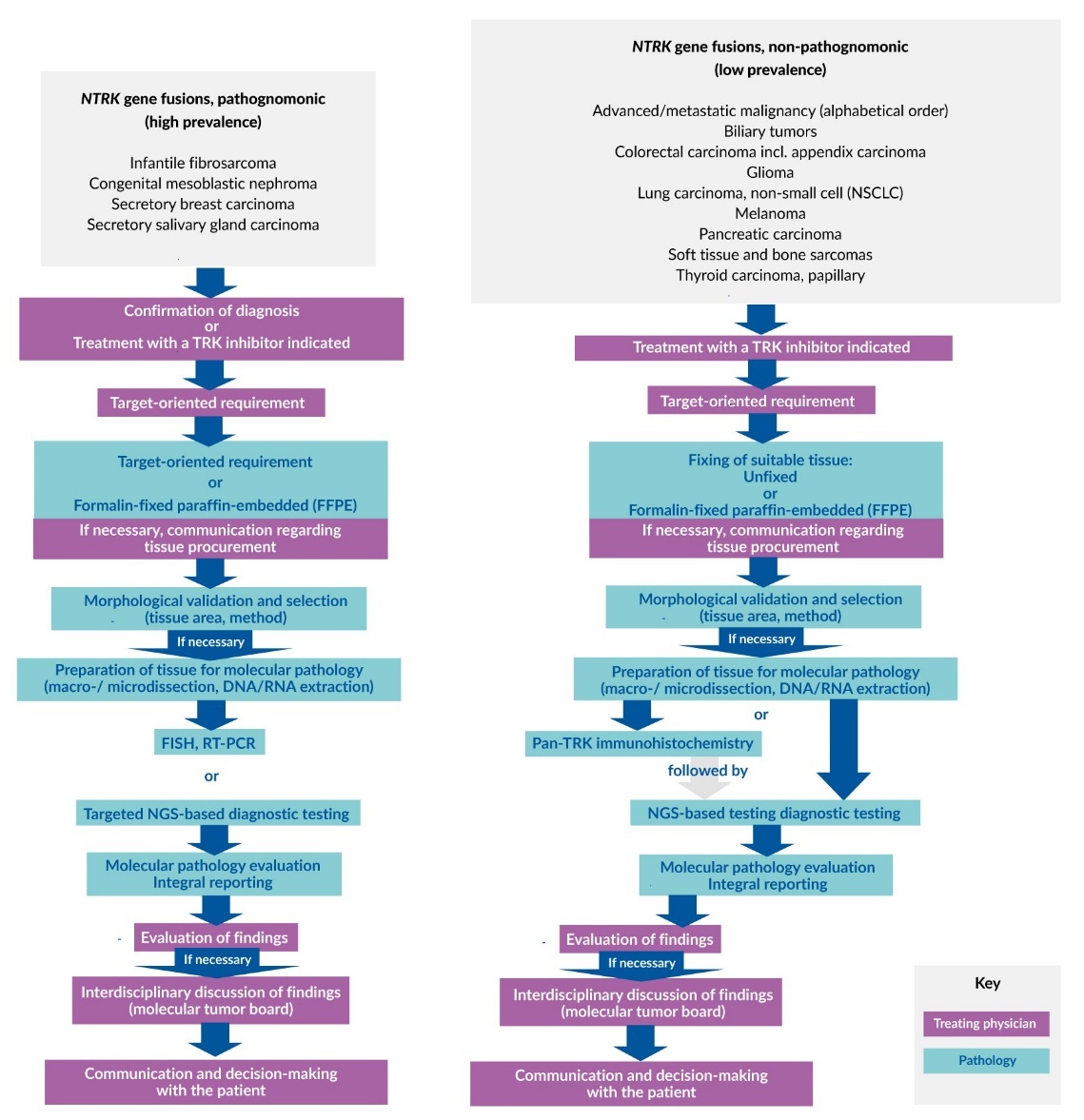
In the algorithm proposed by the DGHO, two different scenarios are considered from a pathological/anatomical viewpoint which take into account tumor types with either common or rare NTRK gene fusions (Figure 2)1:
-
Diagnostic NTRK testing: This may be utilized if the fused NTRK genes are pathognomonic, i.e. in rare tumor types that display a high prevalence of fusions. If the presence of such a tumor is suspected based on histology, the initial focus will be on confirming the diagnosis by means of NTRK testing. Following a positive finding for NTRK gene fusion, patients may then benefit from subsequent treatment with a TRK inhibitor. Diagnostic NTRK testing is particularly important not only for rare tumors such as infantile fibrosarcoma, congenital mesoblastic nephroma, secretory breast carcinoma, and secretory salivary gland carcinoma, but also for high-grade gliomas in children (<3 years) and pediatric papillary thyroid carcinoma.1,16,18,21 Notably, it is strongly recommended to test for NTRK gene fusion in rare tumors as therapy may be used in early lines due to the lack of treatment options.
-
Theragnostic NTRK testing: This may be considered if gene fusion detection is not pathognomonic but necessary for the initiation of targeted TRK inhibitor therapy. This form of NTRK testing is indicated especially for more common tumors such as colon carcinoma, lung carcinoma, and soft tissue sarcoma in adults in which NTRK gene fusion, although rare, is of great therapeutic relevance to patients in the event of a positive test result.1
ALGORITHM FOR NTRK TESTING DEVELOPED BY SOLOMON ET AL.
Solomon et al.21 propose an algorithm that also distinguishes between tumor types that commonly and rarely harbor NTRK gene fusions, with histology- or genomic-based triaging being carried out depending on tumor type (Figure 3).21 Histology-based triages are recommended for diagnostic NTRK testing (see above) by immunohistochemistry for secretory carcinomas and by FISH or RNA-based NGS panel for sarcomas. Genomic based triages are applicable for the use of targeted therapies and are of relevance for all tumors which very rarely harbor NTRK gene fusions, such as most carcinomas, but also gliomas in adults and melanomas.21 In settings where broad up-front testing is not possible, genomic-based triaging provides a means of identifying patients with a higher probability of harboring an NTRK gene fusion, so that screening can then be carried out in a targeted manner. In the case of lung carcinoma, these are patients lacking other driver mutations and generally with a low tumor mutational burden. In patients with colorectal carcinoma, lack of tumor drivers (KRAS, BRAF) and high sporadic microsatellite instability are important criteria. In other tumors (e.g., carcinomas, gliomas, and melanomas) there is also an increased probability of NTRK gene fusion in cases lacking of driver mutations.21 For sarcomas, comprehensive fusion tests are again increasingly being performed in the primary diagnostic evaluation without ‒ as with a carcinoma ‒ waiting for the results of DNA-based triaging.21 In these cases, NTRK fusions would be detected systematically.
ALGORITHM FOR PREDICTIVE TESTS IN ADVANCED NSCLC
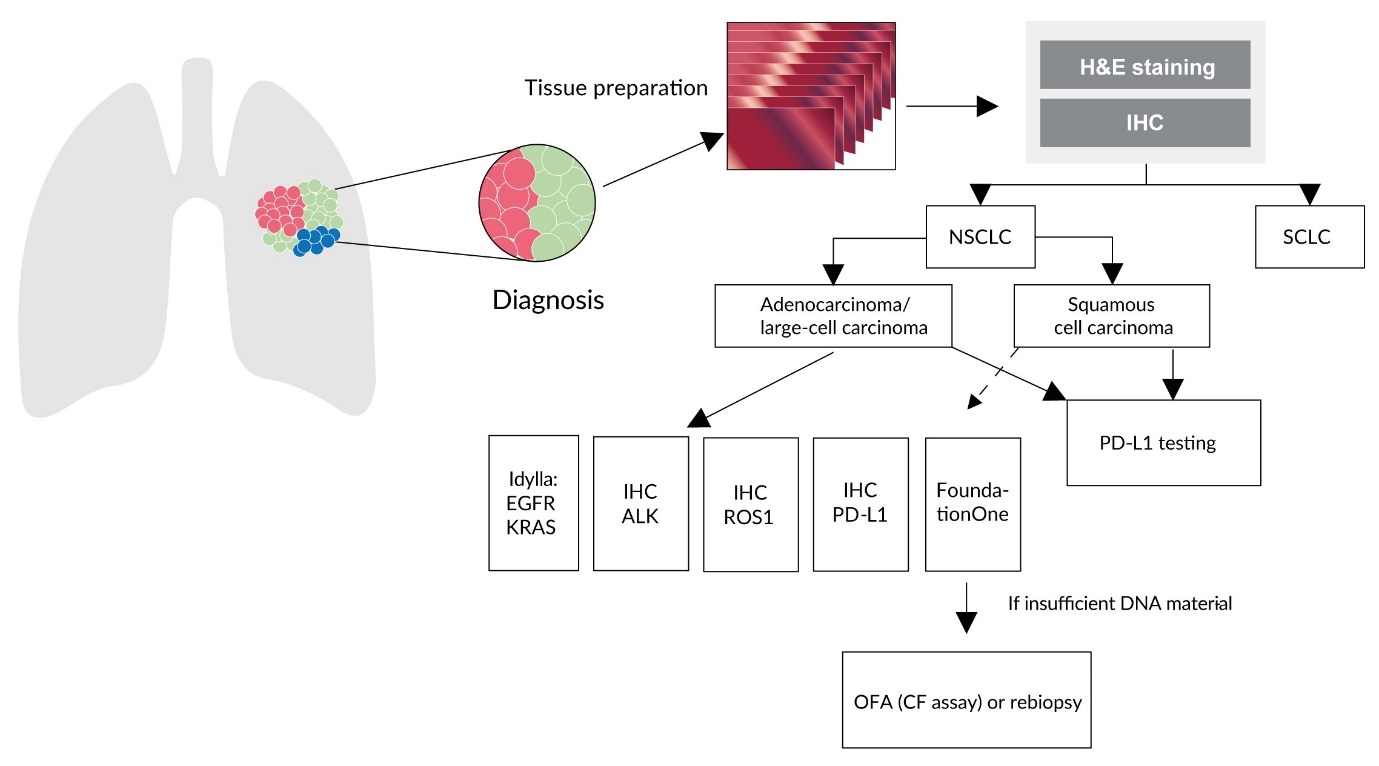
The algorithm used at University Hospital Zurich is illustrated in Figure 4 – based on the diagnosis of a metastatic non-small cell lung cancer (NSCLC) patient.26 The first step is differentiation into small cell (SCLC) and NSCLC, with the latter further divided into adenocarcinoma/not otherwise specified (NSCLC-NOS) and squamous cell carcinoma.26 Testing can then be carried out by immunohistochemistry for ALK, ROS1, PDL-1 and using the real-time PCR-based Idylla™ platform for EGFR and KRAS.26 The NGS-based FoundationOne® CDx assay can be used to produce a complete molecular tumor profile, which may enable, for example, NTRK1, NTRK2, NTRK3, and corresponding fusion genes to be detected.26 FoundationOne CDx test is performed in Switzerland at the Department of Pathology and Molecular Pathology of the University Hospital in Zurich. This test is already being used in routine clinical practice, allowing identification of NTRK alterations among many other genes. If there is insufficient DNA material for such a FOneCDx analysis, the NGS-based Oncomine™ Focus Assay or Oncomine™ Comprehensive Assay can be used instead.26 At the University Hospitals of Basel and Lausanne, a similar algorithm is used that relies primarily on fusion transcript detection using the RNA-based Archer FusionFlex NGS panel.27 In Geneva, the Illumina pan-cancer fusion panel is used. In Locarno, the Archer panel is currently used only if requested by an oncologist. However, there are plans to carry out an Archer test for all NSCLC adenocarcinomas in the coming months instead of IHC/FISH for ALK-ROS-RET.
As with NSCLC, a different strategy should be devised for each tumor type, based on the available data and recommendations, including ESMO recommendations and those presented above.
CLINICAL AND ECONOMIC ASPECTS
Predictive testing to identify patients who may benefit from larotrectinib and other TRK-inhibitors is necessary from a clinical and economic viewpoint. In colorectal carcinoma for example, high microsatellite instability (MSI-h) status is predictive of response to checkpoint inhibitors (CPI) and is already being tested in most larger pathology departments in Switzerland. Notably, physicians/hospitals outside a University Hospital already have access to NTRK testing via the network and interaction to a molecular tumor board provided by different pathology departments. Future health economic assessments of NTRK testing strategies to identify the most cost-effective approach and economic impact of such strategies is required. Indeed, a Swiss cost-effectiveness analysis of different predictive testing strategies in the adjuvant setting has already been performed for breast cancer testing.28
CONCLUSIONS
Because the presence of NTRK gene fusions opens up new treatment options, it is necessary to ensure that diagnostic testing in pathology institutes is of a high quality and takes place within an accredited framework. Detection can be carried out by means of immunohistochemistry, next-generation sequencing or fluorescence in situ hybridization. Various algorithms for NTRK testing are proposed by international organizations. In Switzerland, the requirement for testing to be carried out promptly, so that appropriate therapies can be initiated as early as possible, is being met by pathology institutes.
TAKE-HOME MESSAGES
-
With the authorization of the first TRK inhibitor in Switzerland, larotrectinib (Vitrakvi®), targeted NTRK testing is gaining importance.
-
NTRK gene fusion detection is indicated for rare pediatric tumors and for adult patients with rare forms of secretory breast cancer or salivary gland cancer.
-
NTRK testing can also open up new treatment options for common metastatic tumors in adults (lung and colon carcinoma).
-
Various diagnostic algorithms exist for detecting NTRK gene fusions, each of which propose a specific approach for defined clinical scenarios.
-
Treatments of generally rare NTRK-positive tumors with a TRK inhibitor should be documented in quality-assured, internationally networked registers.1
COI
The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
All authors contributed to and approved the final manuscript.
ACKNOWLEDGEMENTS
We should like to thank Dr. Ellen Heitlinger (H+O communications Ltd., Zurich, Switzerland) for her support with editing the manuscript (including fact checking, linguistic editing, referencing, creation of figures, formatting, proofreading), supported financially by Bayer (Schweiz) AG. Bayer (Schweiz) AG was involved in the final fact check before submission, although the ultimate responsibility for the contents, conclusions and interpretations lies solely with the authors.



_for_detecting_*ntrk*_gene_fusions.png)


_for_detecting_*ntrk*_gene_fusions.png)